Science Spyglass
Why is shRNA better than siRNA for gene knock down in screening campaigns?
One of the first challenges to face in the drug discovery process, is the generation of reliable in-vitro models that mimic the pathological conditions of the disease being studied. Many diseases are caused by the loss of function of a gene, with consequences on downstream pathways and detrimental effects on cell functions. Different approaches can be used in-vitro to obtain a loss of function model at the DNA, RNA or protein level.
We are not new to the use of RNA interference for silencing the mRNA transcripts of such genes, having worked for many years with small interfering RNA (siRNA) transfection. Despite allowing for a great flexibility and ease of use, this approach poses some issues in terms of throughput and variability, when screening large compound libraries to find molecules able to revert the pathological conditions into healthy ones, mainly due to the need to transfect many batches of cells during the screening process.
An alternative to siRNA transfection, well-suited for high-throughput screening (HTS) and hit-to-lead campaigns, is the development of assays based on inducible short hairpin RNA (shRNA) expression, to knock down the gene of interest.

How does it work?
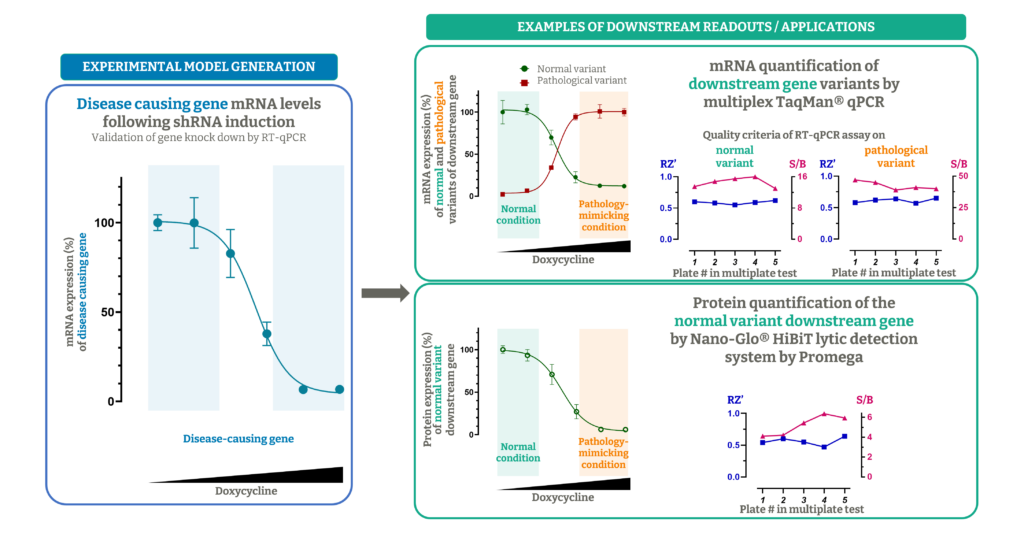
In our experimental model, we treat the cells with lentiviral particles encoding for inducible shRNA directed against the gene of interest, in order to generate a stable cell line able to mimic the pathological condition, upon gene downregulation triggered by doxycycline.
What are the advantages?
Compared to siRNA transfection, that is a transient approach, the shRNA viral transduction generates a stable cell line, and the gene downregulation can be sustained over time by treatment with doxycycline, making this approach very suitable for HTS campaigns and hit-to-lead programs. Indeed, the entire process is particularly fit for automation as it requires fewer handling steps.
One additional important advantage of this approach is that the shRNA induction with doxycycline is dose- dependent (the possibility to modulate the expression is especially important when studying essential and lethal genes) and time-dependent (reversible upon withdrawal of doxycycline).
When to apply this approach?
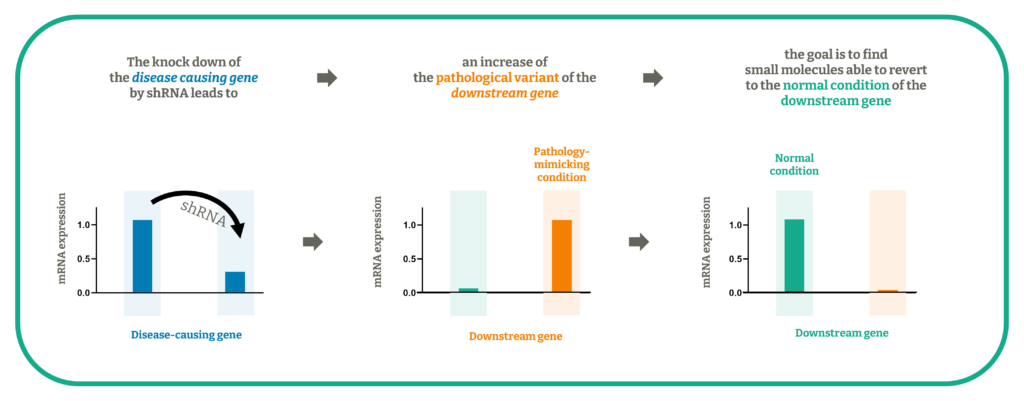
Independently of the therapeutic area, this approach is particularly meaningful when studying diseases characterized by a depletion or mutation of a “disease causing gene” (loss of function) with the final aim of identifying small molecules that revert such pathological conditions to the normal state. The experimental disease-related model can be used also to study “downstream genes” regulated or at least influenced by the disease causing gene that will be knock down by specific shRNA.
A case study:
focus on a disease caused by the mis-splicing of a target gene
We developed a disease model transducing shRNA expressing lentivirus into a human neuroblastoma cell line, to silence a splicing factor (the disease-causing gene), whose downregulation causes a mis-splicing in a target gene (downstream gene).
Related content



