SARS-CoV-2 Viral Assays
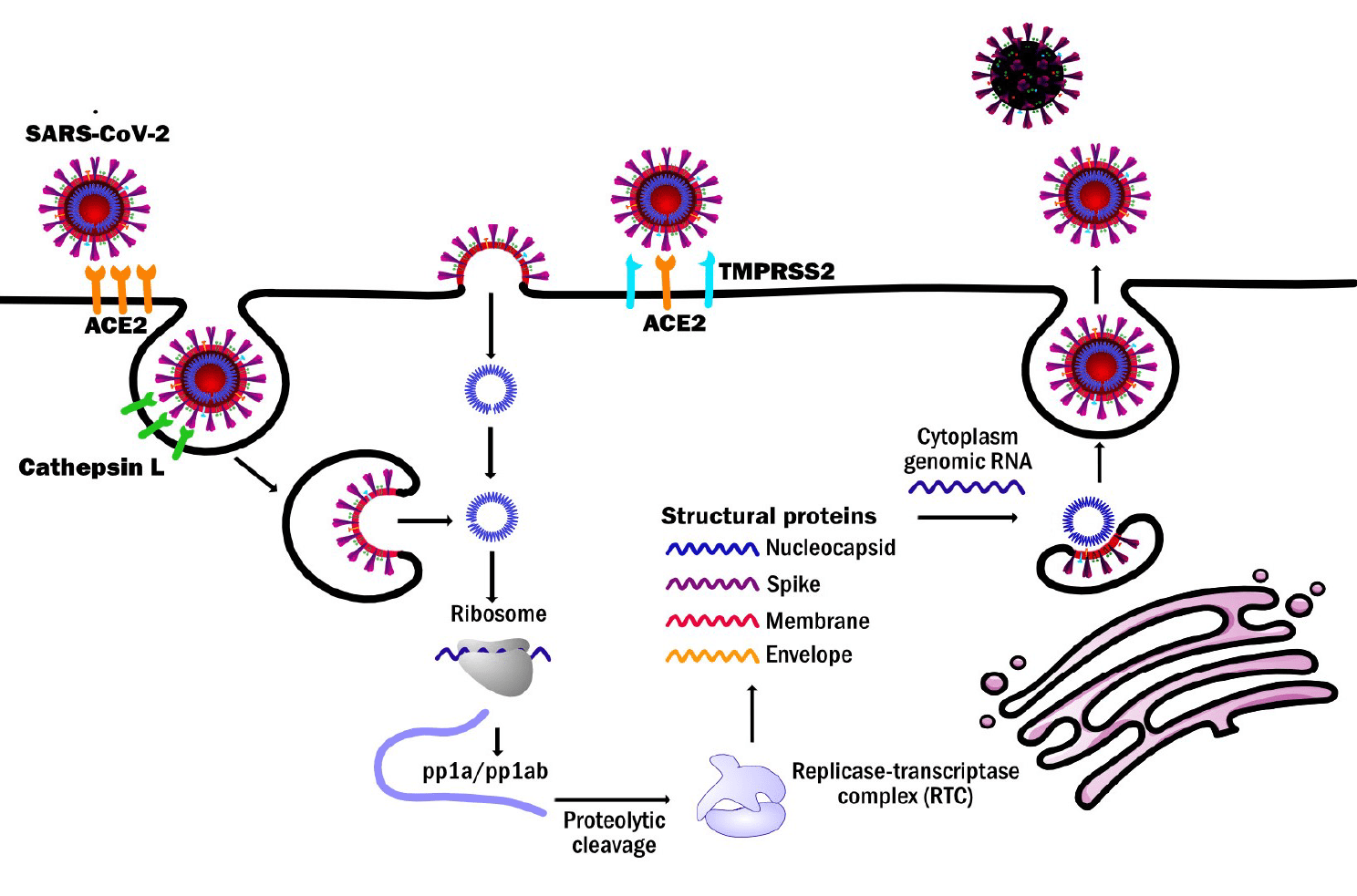
Being the first stage in infection, entry of SARS-CoV-2 into host cells is an extremely attractive therapeutic intervention point. Two cell-based SARS-CoV-2 viral entry assays involving the use of pseudotyped virus-like particles (VLPs) have been developed and optimized:
- SARS-CoV-2 HiBiT VLP viral entry assay
- SARS-CoV-2 reporter-based VLP viral entry assay
These assays can be adapted to cover the most known variants (e.g., alpha, delta and omicron) of the SARS-CoV-2 virus.
Note on VLPs: Virus-like particles (VLPs) are stable, non-replicating nanostructures comprised of viral structural proteins and a lipid envelope. They do not contain viral genome. For developing our assay we use VLPs that express the SARS-CoV-2 spike protein on the surface, thus retaining the ability to mimic the infectious process of the SARS-CoV-2 virus. VLPs are safe and therefore do not require handling in high containment BSL3 facilities.
The approach based on VPLs may also potentially be applied to develop assays tackling a wide range of other viruses.
This viral entry assay has been optimized for HTS application in collaboration with Promega Corporation to identify inhibitors of the viral attachment, as well as the fusion and uncoating steps of the viral replicative cycle. The assay relies on Promega’s NanoBiT® technology and is based on the reconstitution of the NanoBiT luciferase, whose small portion (HiBiT) is tagged on VLPs and large portion (LgBiT) is expressed by the host target cells (as well as ACE2 and TMPRSS2, to allow viral entry).
- Assay developed with HiBiT-tagged VLPs and detection substrate (both Promega) in combination with a cell line developed by Axxam and validated with reference inhibitor compounds and neutralizing monoclonal antibodies, also against different SARS-CoV-2 spike variants
- 384 well plate format
- Fast luminescent signal (FLIPRTETRA) upon viral entry (3 hours or less)
- Optimized assay (fast, highly sensitive) for compound profiling and compatible with HTS applications
- Specificity assay available (based on VSV-G pseudotyped VLPs)
- Possible real-time assessment of viral entry
This viral entry assay allows the identification of inhibitors of SARS-CoV-2 viral entry and it’s based on the interaction between the SARS-CoV-2 Spike protein on the surface of VLPs and a target cell line developed at Axxam expressing ACE2 and TMPRSS2, which have a key role in the viral entry step. The VLPs are able to transduce into the target cell a transfer vector, expressing two reporter genes (GFP, Nanoluc luciferase), that allow to monitor the viral uptake into the host cells.
- Assays developed with custom-made VLPs, validated for viral entry inhibition with reference serine protease inhibitors (vs TMPRSS2) as well as recombinant ACE2 (ACE2-Fc), which is able to efficiently displace ACE2-Spike binding, thus inhibiting SARS-CoV-2 viral entry
- 384 well plate format
- Read-outs with luminescence (FLIPR) or fluorescence (Operetta)
- Optimized assay for compound profiling (DMSO sensitivity, stability and reproducibility of pharmacology, multiplate test)
- Specificity assay available (based on VSV-G pseudotyped VLPs)
- May be adapted as a neutralizing antibody assay
Three biochemical SARS-CoV-2 assays have been developed and optimized for the identification or repurposing of small molecule inhibitors. Even the biochemical assay tackles the first step of the viral entry (TMPRSS2 and ACE2 assay) but also a key step of viral replication (Mpro assay).
- TMPRSS2 viral entry assay
- ACE2 spike S1 interaction assay
- Mpro inhibition of viral life cycle
The TMPRSS2 viral assay was devised to identify possible inhibitors of the proteolytic activity of TMPRSS2 enzyme. This enzyme is located on the cell surface of human cells and, cleaving Spike protein, helps the viral entry.
- Fluorescence-based enzymatic assay (FRET) employing a surrogate fluorogenic substrate
- 384 well plate format
- Fully optimized working concentrations of hTMPRSS2 and substrate
- Tolerance to DMSO
- Assay validated with reference inhibitor
- Interference assay available
The aim of the ACE/Spike S1 interaction assay is the identification of compounds disrupting the binding occurring between human ACE2 enzyme and viral Spike S1 thus, preventing the viral entry.
- TR-FRET biochemical interaction assay based on commercial tagged proteins
- 384 well plate format
- Fully optimized working concentrations of hACE2 and SARS-CoV2-Spike S1
- Tolerance to DMSO
- Assay validated with reference tools
- Interference assay available
- Fluorescence based assay (FRET) employing a surrogate fluorogenic substrate
- Fully optimized working concentrations of Mpro and substrate
- Functional SARS-CoV-2 Mpro activity assay in 384 and 1536 well plate formats
- Functional SARS-CoV-2 Mpro interference assay
- Functional Mpro orthologous enzymatic assay also for MERS, EV-D68, SARS-CoV-1
- All assays are validated by commercial reference inhibitors
- Validated for use with HTS automation